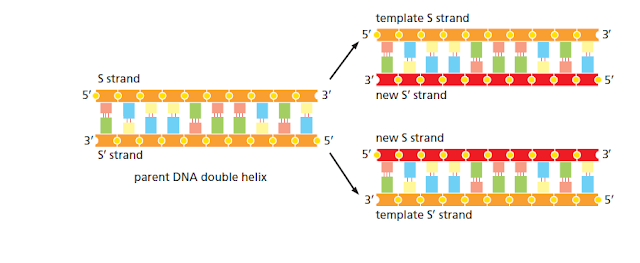
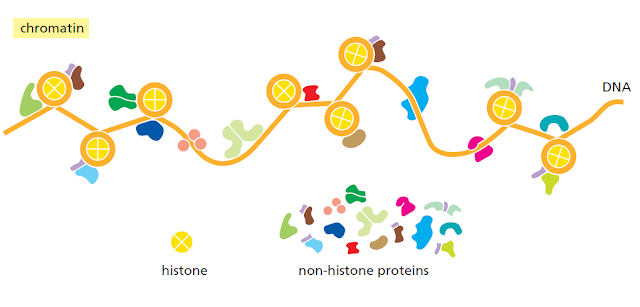
RNA to Protein : Degradation and Splicing

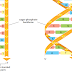
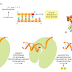
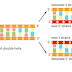
Molecule is being transported from the nucleus to the cytosol. As its 5ʹ end emerges
from a nuclear pore, the mRNA is met by a ribosome, which begins to translate
it. As translation proceeds, the exon junction complexes (EJCs) that are bound
to the mRNA at each splice site are displaced by the moving ribosome. The normal
stop codon will lie within the last exon, so by the time the ribosome reaches
it and stalls, no more EJCs will be bound to the mRNA. In this case, the mRNA
“passes inspection” and is released to the cytosol where it can be translated in earnest
(Figure 6–76). However, if the ribosome reaches a stop codon earlier, when
EJCs remain bound, the mRNA molecule is rapidly degraded. In this way, the first
round of translation allows the cell to test the fitness of each mRNA molecule as it
exits the nucleus.
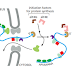
Nonsense-mediated decay may have been especially important in evolution,
allowing eukaryotic cells to more easily explore new genes formed by DNA rearrangements,
mutations, or alternative patterns of splicing—by selecting only those
mRNAs for translation that can produce a full-length protein. Nonsense-mediated
decay is also important in cells of the developing immune system, where
the extensive DNA rearrangements that occur.
.png)