DNA to RNA : RNA Splicing
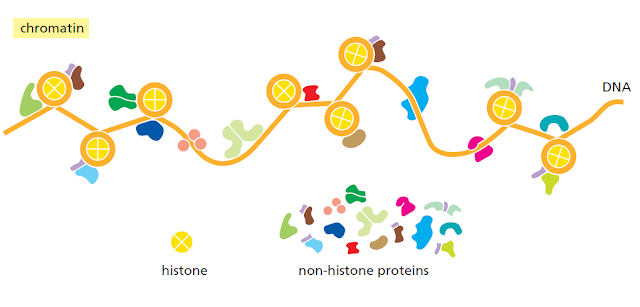
Although it may seem at first counterintuitive, the way a gene is packaged into
chromatin can affect how the RNA transcript of that gene is ultimately spliced.
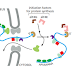
Nucleosomes tend to be positioned over exons (which are, on average, close to the
length of DNA in a nucleosome), and it has been proposed that these act as “speed
bumps,” allowing the proteins responsible for exon definition to assemble on the
RNA as it emerges from the polymerase. In addition, changes in chromatin structure
are used to alter splicing patterns. There are two ways this can happen. First,
because splicing and transcription are coupled, the rate at which RNA polymerase
moves along DNA can affect RNA splicing. For example, if polymerase is moving
slowly, exon skipping (see Figure 6–30A) is minimized: assembly of the initial
spliceosome may be complete before an alternative choice of splice site even
emerges from the RNA polymerase. The nucleosomes in condensed chromatin
can cause polymerase to pause; the pattern of pauses in turn affects the extent of
RNA exposed at any given time to the splicing machinery.
RNA Splicing Shows Remarkable Plasticity
We have seen that the choice of splice sites depends on such features of the premRNAtranscript as the strength of the three signals on the RNA (the 5ʹ and 3ʹ splice
junctions and the branch point) for the splicing machinery, the co-transcriptional
assembly of the spliceosome, chromatin structure, and the “bookkeeping” that
underlies exon definition. We do not know exactly how accurate splicing normally
is because, as we see later, there are several quality control systems that rapidly
destroy mRNAs whose splicing goes awry. However, we do know that, compared
with other steps in gene expression, splicing is unusually flexible.
.png)






0 komentar: