DNA Replication : The Mehcanism

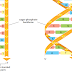
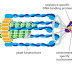
DNA polymerase performs the first proofreading step just before a new nucleotide
is covalently added to the growing chain. Our knowledge of this mechanism
comes from studies of several different DNA polymerases, including one produced
by a bacterial virus, T7, that replicates inside E. coli. The correct nucleotide
has a higher affinity for the moving polymerase than does the incorrect nucleotide,
because the correct pairing is more energetically favorable. Moreover, after
nucleotide binding, but before the nucleotide is covalently added to the growing
chain, the enzyme must undergo a conformational change in which its “grip”
tightens around the active site (see Figure 5–4). Because this change occurs more
readily with correct than incorrect base-pairing, it allows the polymerase to “double-
check” the exact base-pair geometry before it catalyzes the addition of the
nucleotide. Incorrectly paired nucleotides are harder to add and therefore more
likely to diffuse away before the polymerase can mistakenly add them.
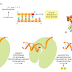
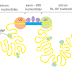
The next error-correcting reaction, known as exonucleolytic proofreading,
takes place immediately after those rare instances in which an incorrect nucleotide
is covalently added to the growing chain. DNA polymerase enzymes are
highly discriminating in the types of DNA chains they will elongate: they require
a previously formed, base-paired 3ʹ-OH end of a primer strand (see Figure 5–4).
Those DNA molecules with a mismatched (improperly base-paired) nucleotide
at the 3ʹ-OH end of the primer strand are not effective as templates because the
polymerase has difficulty extending such a strand. DNA polymerase molecules
correct such a mismatched primer strand by means of a separate catalytic site
(either in a separate subunit or in a separate domain of the polymerase molecule,
depending on the polymerase). This 3ʹ-to-5ʹ proofreading exonuclease clips off any
unpaired or mispaired residues at the primer terminus, continuing until enough
nucleotides have been removed to regenerate a correctly base-paired 3ʹ-OH terminus
that can prime DNA synthesis. In this way, DNA polymerase functions as a
“self-correcting” enzyme that removes its own polymerization errors as it moves
along the DNA (Figure 5–8 and Figure 5–9).
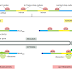
The self-correcting properties of the DNA polymerase depend on its requirement
for a perfectly base-paired primer terminus, and it is apparently not possible
for such an enzyme to start synthesis de novo, without an existing primer.
By contrast, the RNA polymerase enzymes involved in gene transcription do not
need such an efficient exonucleolytic proofreading mechanism: errors in making
RNA are not passed on to the next generation, and the occasional defective RNA
molecule that is produced has no long-term significance. RNA polymerases are
thus able to start new polynucleotide chains without a primer.

.png)















0 komentar: